Coin Laboratory: Genomics and bioinformatics in infectious disease and cancer
-
Head of Laboratory

Professor Lachlan Coin+61383443831
Research Overview
View a comprehensive listing of Professor Coin's research publications here.
The Coin Group develops genomic and transcriptomic tools to develop biomarkers for rapid characterization of disease state and prediction of drug susceptibility, with the aim of decreasing the time taken from hospital admission to administering the right treatment. We also develop biomarkers for measuring treatment response. We currently focus on infectious disease as well as cancer.
We utilize approaches from high-dimensional statistics, information theory and machine learning, including deep neural networks. We aim to implement streaming algorithms, which process data as soon as it is generated, providing real-time inference and visualization of both the most likely predicted disease state, as well as uncertainty in those predictions, which decrease as more data is collected.
Much of our current research utilizes real-time nanopore sequencing, based on its unique ability to generate sequence data in real-time, as well as its capability to sequence native DNA and RNA. We have emerging interests in single-cell long-read RNA and DNA sequencing, particularly as they relate to improving diagnostic and prognostic tools.
In infectious disease, we collaborate with chemists and materials scientists to develop techniques for isolating bacteria from complex biological matrices. We apply these approaches in conjunction with real-time nanopore sequencing to sequence bacterial genomes directly from clinical samples.
We have ongoing interests in evolution of complex regions of the genome, particularly highly repetitive regions. We are currently working utilizing long nanopore reads to characterize complex regions of tumour genomes inaccessible to short-read sequencing technologies. We are interested in the role in which complex rearrangements in these repetitive regions can influence phenotype, including drug resistance. We have also worked on using circulating tumour DNA to non-invasively characterise copy number variation in primary tumours.
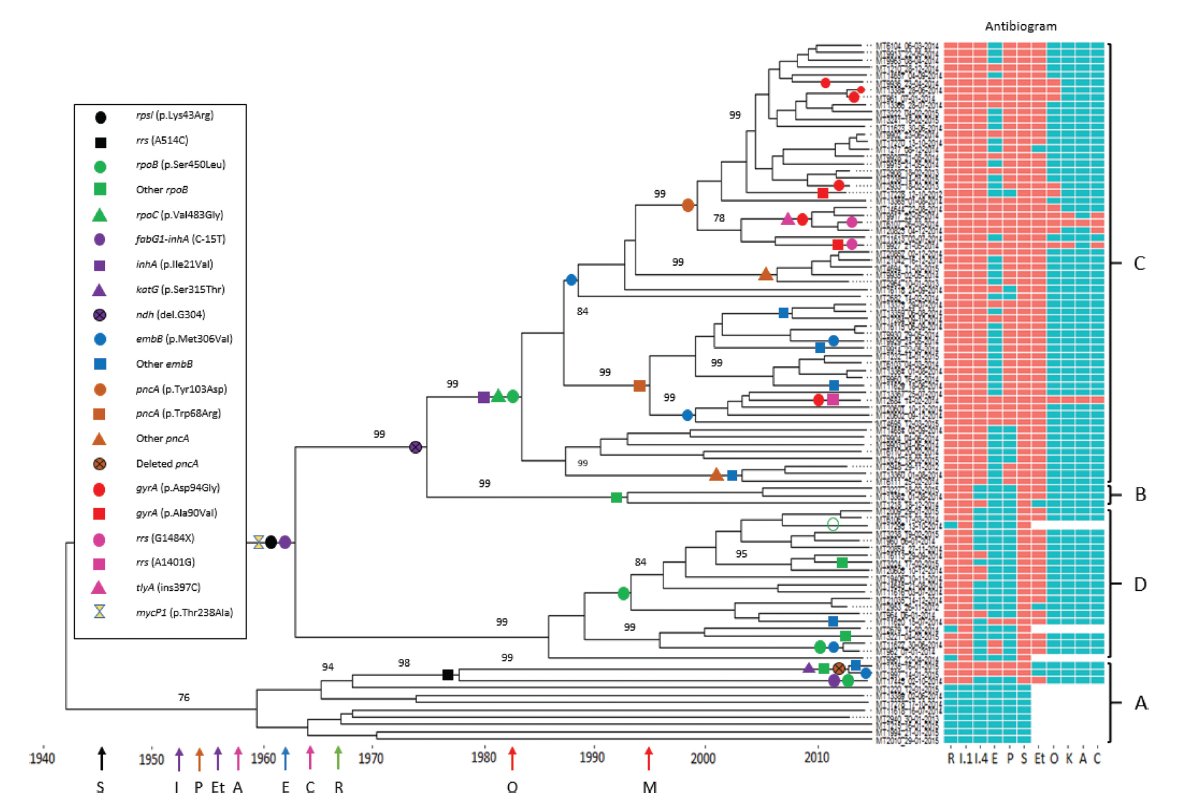
Image 1: Phylogeny of Mycobacterium tuberculosis strains in Papua New Guinea showing estimated dates of acquisition of drug resistance mutations.
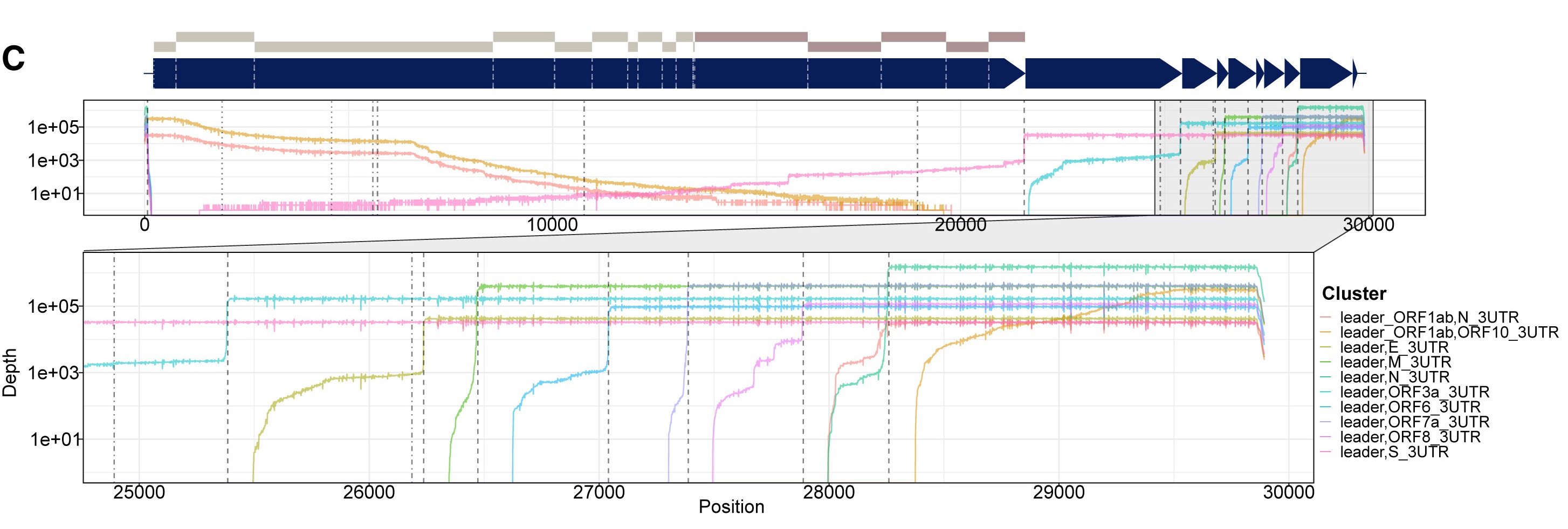
Image 2: Depth of coverage of transcripts in SARS-Cov2 revealed by long read nanopore sequencing.
Staff
Professor Lachlan Coin, Group leader
Dr Miranda Pitt, Research Officer
Dr Fuyi Li, Research Officer
Dr Eike Steinig, Research Officer
Jessie Chang, PhD Student
Daniel Rawlinson, PhD Student
Collaborators
Professor Tim Stinear, Department of Microbiology and Immunology, The University of Melbourne
Professor Deborah Williamson, Department of Microbiology and Immunology, The University of Melbourne
Professor Fred Hollande, The University of Melbourne Centre for Cancer Research
Professor Matt Cooper, University of Queensland
Associate Professor Mark Blaskovich, University of Queensland
Associate Professor Luregn Schlapbach, University of Queensland
Dr Seweryn Bialasiewicz, University of Queensland
Dr Claire Wainwright, University of Queensland
Professor Mike Levin, Imperial College London
Dr Myrsini Kaforou, Imperial College London
Dr Ulrich Van Both, Ludwig Maximilian University of Munich
Research Opportunities
This research project is available to PhD students, Masters by Research, Honours students, Post Doctor Researchers to join as part of their thesis.
Please contact the Research Group Leader to discuss your options.
Research Outcomes
Brian Eley, Lachlan Coin, Michael Levin, Suzanne Anderson, Method of detecting active tuberculosis in children in the presence of a co-morbidity, WO2014067943A1
Research Publications
Teng, H., Cao, M. D., Hall, M. B., Duarte, T., Wang, S., & Coin, L. J. (2018). Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning. GigaScience, 7(5), giy037.
Zhou, C., Olukolu, B., Gemenet, D. C., Wu, S., Gruneberg, W., Cao, M. D., ... & Coin, L. J. (2020). Assembly of whole-chromosome pseudomolecules for polyploid plant genomes using outbred mapping populations. Nature Genetics, 52(11), 1256- 1264.
Chang, J. J. Y., Rawlinson, D., Pitt, M. E., Taiaroa, G., Gleeson, J., Zhou, C., ... & Coin, L. J. (2021). Transcriptional and epi-transcriptional dynamics of SARS-CoV-2 during cellular infection. Cell Reports, 35(6), 109108.
Nguyen, S. H., Cao, M. D., & Coin, L. J. (2021). Real-time resolution of short-read assembly graph using ONT long reads. PLOS Computational Biology, 17(1), e1008586.
Cao, M. D., Nguyen, S. H., Ganesamoorthy, D., Elliott, A. G., Cooper, M. A., & Coin, L. J. (2017). Scaffolding and completing genome assemblies in real-time with nanopore sequencing. Nature communications, 8(1), 1-10.
Pitt, M. E., Elliott, A. G., Cao, M. D., Ganesamoorthy, D., Karaiskos, I., Giamarellou, H., ... & Coin, L. J. (2018). Multifactorial chromosomal variants regulate polymyxin resistance in extensively drug-resistant Klebsiella pneumoniae. Microbial genomics, 4(3).
Bainomugisa, A., Duarte, T., Lavu, E., Pandey, S., Coulter, C., Marais, B. J., & Coin, L. M. (2018). A complete high-quality MinION nanopore assembly of an extensively drug-resistant Mycobacterium tuberculosis Beijing lineage strain identifies novel variation in repetitive PE/PPE gene regions. Microbial genomics, 4(7).
Research Projects
For project inquiries, contact our research group head.
Faculty Research Themes
School Research Themes
Infection & Immunity, Molecular Mechanisms of Disease
Key Contact
For further information about this research, please contact Head of Laboratory Professor Lachlan Coin
Department / Centre
Unit / Centre
Coin Laboratory: Genomics and bioinformatics in infectious disease and cancer
MDHS Research library
Explore by researcher, school, project or topic.