Paul Gooley laboratory
-
Head of Laboratory

Professor Paul Gooley+61 0437 835 059 (mobile)
+61 3 8344 2273 (office)
Research Overview
Research in the Gooley laboratory uses solution structural biology techniques to understand the mechanism of action of proteins. The major structural tool used is solution nuclear magnetic resonance spectroscopy, but we take advantage of other approaches including isothermal titration calorimetry, surface plasmon resonance, X-ray scattering, X-ray crystallography and fluorescence spectroscopy. Research focuses on protein-protein and protein-ligand interactions that are linked to disease with the goal of finding novel ligands or designing new therapies for treatment.
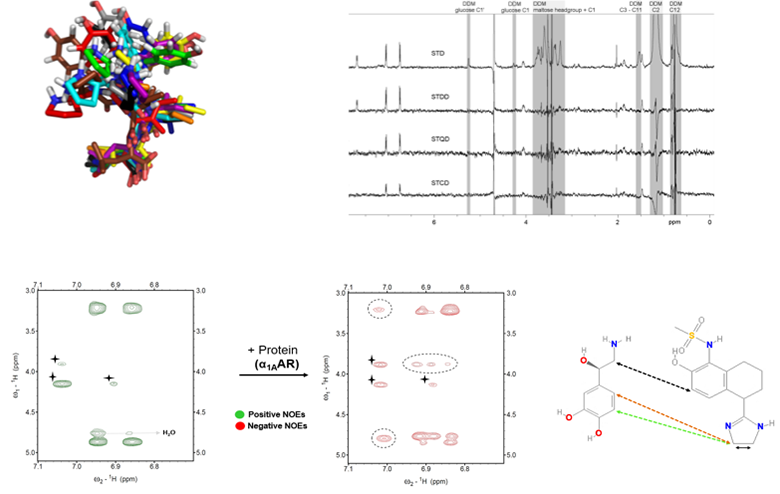
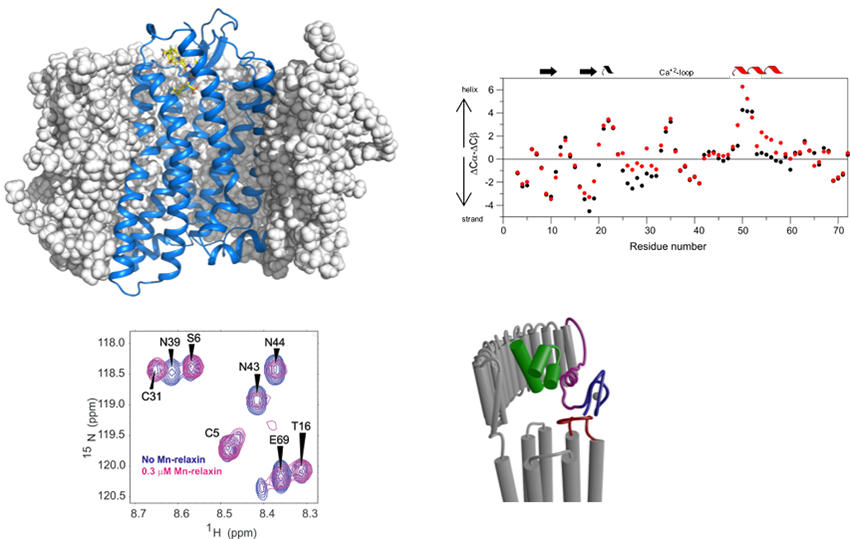
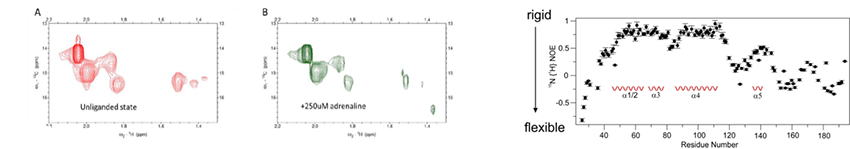
A major project is the investigation of how G-protein coupled receptors transmit biological signals. This program focuses on the peptide receptors RXFP1 (the receptor for the hormone relaxin) and NTS1 (the receptor for neurotensin) and the two aminergic receptors, alpha1A and alpha1B adrenoreceptors. RXFP1 is a multidomain receptor and we have made significant progress in understanding the roles of different regions of the protein in activation. The two adrenoreceptors have apparent opposing roles and so subtype selectivity is essential to minimize side effects. Understanding mechanism and ligand binding are important for these objectives.
Another area of research is studying the processes of cell evasion and replication of viruses. We have focused on the P protein of lyssavirues (including the rabies virus) which has essential roles in evasion and replication. P protein by interating with the host's transcription factors, the STAT proteins, antagonizes the interferon-mediated pathway. The P protein is an essential effector of relication by interacting with both the viral N and L proteins. As substantial regions of P protein are intrinsically disordered we want to learn the importance of the structural plasticity in these processes.
Staff
Fei Yan, Research Assistant
Ashish Sethi, Postdoc
Chris Armstrong, Postdoc
Fengjie Wu, PhD student, co-supervised with Dan Scott (Florey)
Tasneem Vaid, PhD student, co-supervised with Dan Scott (Florey)
Alamgir Hossain, PhD student
Jingyu Zhan, PhD student
Samantha Lagaida, PhD student
Shoni Bruell, PhD student, co-supervised with Ros Bathgate (Florey)
Winnie Wu, PhD student, co-supervised with Ross Bathgate (Florey)
Mohammad Tanipour, PhD student, co-supervised with Dan Scott (Florey)
Joey Ibrahim , Honours student
Siqiong Zhen, Masters student
Collaborators
Professor Ross Bathgate (Florey
Dr Dan Scott (Florey)
Dr Mike Griffin
Professor Spencer Williams
Dr Akhter Hossain (Florey)
Dr David Thal (Monash)
Dr David Chalmers (Monash)
Associate Professor Martin Scanlon (Monash)
Dr Greg Moseley (Monash)
Dr Natalie Borg (Monash)
Dr Margaret Johnson (Alabama, Birmingham)
Dr Henry Butt (Bioscreen, Yarraville, Melbourne)
Funding
NHMRC project grant: “Understanding the mechanism of GPCR activation to enable rational drug discovery”
NHMRC project grant: “Unravelling the binding and activation mechanism of a complex G protein-coupled receptor”
NHMRC project grant: “Defining the Molecular Mechanisms of Lyssavirus Replication and Immune Evasion: the P protein Axis”
Research Publications
Click here for the results of a PubMed search of Paul's publications.
Click here for the results of a Google Scholar analysis of Paul's publications.
Research Projects
For project inquiries, contact our research group head.
Faculty Research Themes
Infection and Immunology, Neuroscience
School Research Themes
Biomedical Neuroscience, Cellular Imaging & Structural Biology, Molecular Mechanisms of Disease, Therapeutics & Translation
Key Contact
For further information about this research, please contact Head of Laboratory Professor Paul Gooley
Department / Centre
Unit / Centre
MDHS Research library
Explore by researcher, school, project or topic.