Structure and function of bacterial "cytomotive" filaments
-
Project Leader

Dr Debnath Ghosal+61 3 8344 8441
Project Details
Until the early 1990s, cytoskeletal proteins were believed to be the hallmarks of eukaryotic cells. However, in the last three decades, the discovery of bacterial homologs of eukaryotic actin (e.g. MreB), tubulin (FtsZ), and intermediate filament (IF) (e.g. crescentin) proteins have dramatically changed our perception. Recent studies have revealed that the catalogue of bacterial cytoskeletal proteins is potentially much longer than previously thought. Bioinformatic analyses of published genomic data have shown that there are at least 35 distinct families of bacterial actins. Similarly, additional homologs of tubulin and IF have been also discovered. Growing evidence suggests that bacteria also harbour additional cytoskeletal filament systems with no eukaryotic homologs such as the Walker A Cytoskeletal ATPases (WACAs), bactofilins, and metabolic enzyme CTP Synthase filaments.
In the ‘absence’ of motor proteins (e.g. kinesins and dyneins), these filamentous systems perform bacterial cell division, generate force, control cell shape, partition DNA and organise intracellular space. One of the key emerging differences between the bacterial and the eukaryotic cytoskeletal systems is that each of the bacterial filaments seem to perform one dedicated function while eukaryotic ones, by virtue of their interaction with a repertoire of adapters and regulatory proteins, perform numerous tasks. How and when this transition happened in evolution is still unclear. We need to investigate many more bacterial filament systems and their interactome to decipher how prokaryotic cytoskeleton was customised for multifunctionality during the evolution of complex eukaryotic cells.
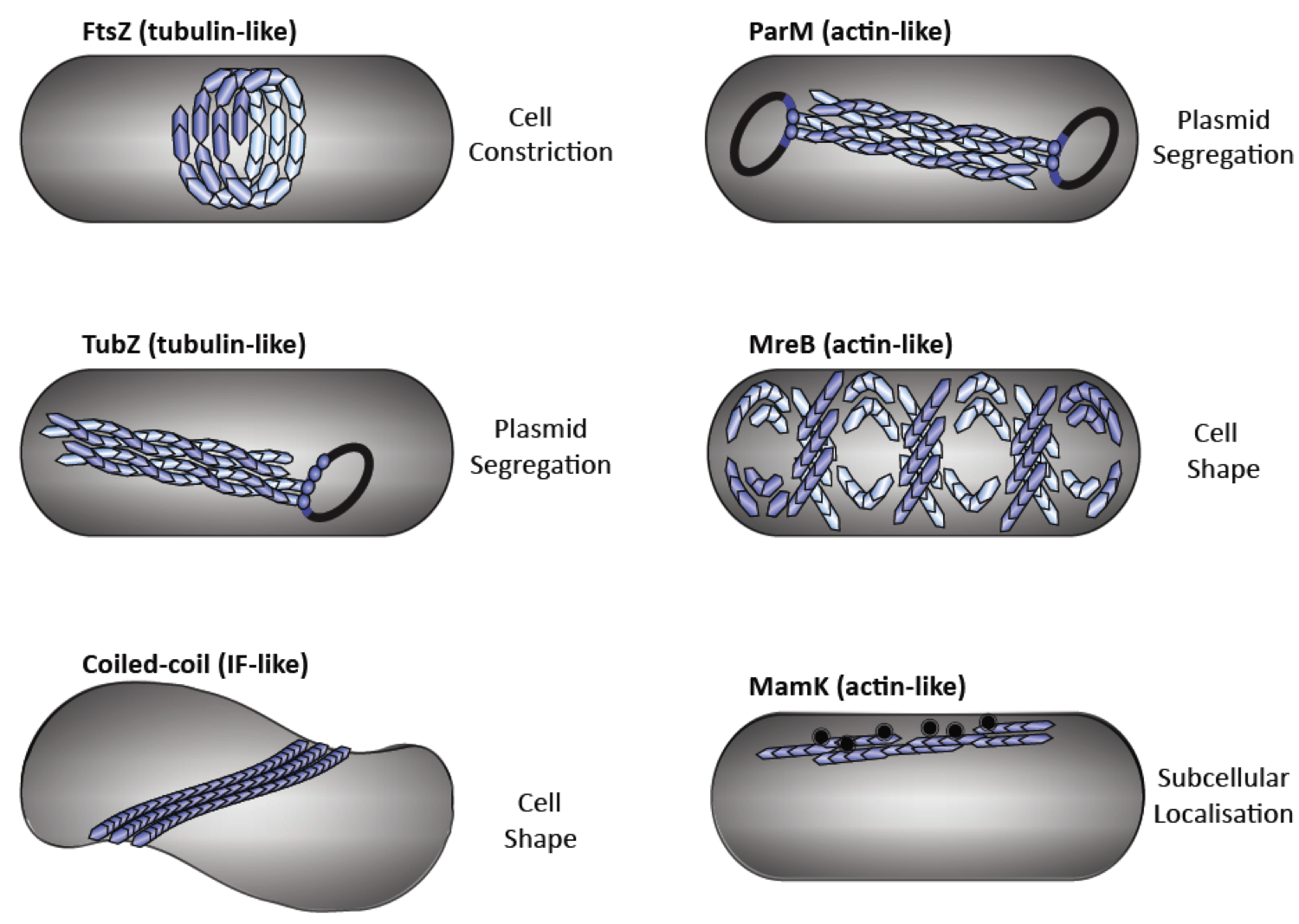
Fig. 1: Diverse functions of cytoskeletal elements in bacteria. Bacterial tubulin homologs: FtsZ performs cytokinesis (a), TubZ executes plasmid segregation (b). Intermediate filament-like coiled-coil proteins (e.g., Crescentin and Bactofilins) regulate cell shape (c). Bacterial actin homologs: ParM executes plasmid segregation (d), MreB maintains cell shape (e) and MamK organises magnetosome chains (f).
Our recent work shows that the bacterial Min proteins (MinC and MinD) negatively regulate the FtsZ-ring (hence divisome) assembly by forming a new class of alternating copolymeric cytoskeletal filaments. These filaments are unique and unrelated to any of the conventional cytoskeletal elements, i.e. actin, tubulin or intermediate filaments and are remotely "related" to eukaryotic septins.
Fig. 2: MinCD co-crystal structure suggests MinCD together form a new class of alternating copolymeric cytomotive filament
We used hybrid methods including, X-ray crystallography, cryo-EM, super-resolution imaging and biochemical reconstitution to demonstrate that the MinCD filaments are essential for proper cell division and confirmed their existence in vivo. We also put forward a mechanistic model for how MinCD filaments might regulate FtsZ-ring assembly in cells.
Research Opportunities
This research project is available to Honours students to join as part of their thesis.
Please contact the Research Group Leader to discuss your options.
Research Group
Faculty Research Themes
School Research Themes
Cellular Imaging & Structural Biology, Molecular Mechanisms of Disease
Key Contact
For further information about this research, please contact the research group leader.
Department / Centre
MDHS Research library
Explore by researcher, school, project or topic.