RNA mediated control of HIV-1 gene expression
-
Project Leader

Professor Damian Purcell+61 3 8344 6753
Project Details
The successful replication of HIV-1 requires intricate RNA processing as well as numerous other mechanisms that control cellular gene expression. Studies by Purcell on the non-productive and productive latent phases of infection showed that the control of gene expression occurs by many events after RNA transcription. These include RNA-capping, transport, splicing, polyadenylation, decay; and chromatin modifications guided by RNA. The events listed impact on the efficiency of RNA transcription initiation as well as elongation and therefore are extremely important in controlling HIV latency.
RNA-mediated Control of HIV latency
Highly effective combination drug therapies slow the development of AIDS, but don't cure an HIV infection, because long-lived persistently infected cells, especially resting memory CD4+ T-cells, harbour integrated HIV DNA that mostly remains silent. The on-going sporadic reactivation of HIV from this latent integrated provirus mandates life-long antiviral therapy and causes incomplete immune recovery and chronic consequences, including cardiovascular and malignant disease. We need to better understand the molecular events governing the silencing or expression from proviral DNA to control latent HIV. This project investigates how HIV enters into and is reactivated from post-integration latency in resting memory CD4+ T-cells. Multiple restrictions prevent viral gene expression from these cells including transcriptional interference, where a strong upstream cellular promoter eclipses the viral promoter, driving transcription of a cellular gene that encases the HIV-1 provirus integrated within an intron. This project is examining how RNA-mediated events, such as the changes to alternative splicing that occur during T-cell maturation, alters the expression of HIV Tat which is a powerful activator of virus production. The project examines cellular and viral factors that impact upon the formation of chimeric cellular-viral mRNAs that include tat exon-2 and the RNA-mediated internal ribosome entry site (IRES) that supports a unique translation-control mechanism that assists in Tat expression and reactivation of productive viral replication. The role of functional small RNAs will also be studied.
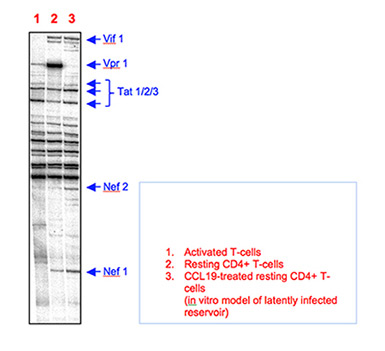
Figure 1: Polyacrylamide urea gel showing differential expression of viral multiply-spliced regulatory mRNAs from primary cells infected with HIV-1NL4.3
Researchers
Con Sonza and Damian Purcell
Research Group
Purcell laboratory: Viral RNA elements, regulation of HIV replication, HIV antibody vaccine
Faculty Research Themes
School Research Themes
Molecular Mechanisms of Disease
Key Contact
For further information about this research, please contact the research group leader.
Department / Centre
MDHS Research library
Explore by researcher, school, project or topic.