Investigating the diversity of the variant antigen genes of Plasmodium falciparum
-
Project Leader

Professor Karen Day+61 3 8344 7644
Project Details
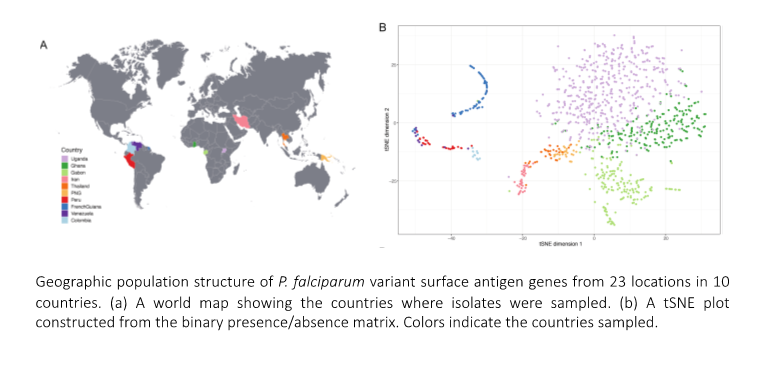
Plasmodium falciparum, the pathogen causing the most virulent form of malaria, continues to present a significant economic and public health burden globally. The pathogen is endemic across many resource poor countries in tropical Africa and parts of Asia, and is re-emerging in Latin America. The pathogen’s success in remaining highly prevalent in many regions can be attributed to the extreme diversity of the major variant surface antigen of the blood stages, known as P. falciparum erythrocyte membrane protein 1 (PfEMP1). This molecule undergoes clonal antigenic variation to switch surface antigens on the infected erythrocyte thereby evading the host immune system and allowing the parasite to persist in the human host for many months. PfEMP1 is encoded by the var multigene family and each parasite possesses up to 60 var genes. Genome sequencing has shown that each parasite carries a different var gene repertoire or haplotype. Bioinformatic analysis of the diversity and population structure of the var genes encoding PfEMP1 is critical for surveillance and prevention of malaria as well as for the design of any variant-specific vaccine targeting PfEMP1 based on an understanding of local herd immunity. For these reasons we are investigating the diversity and geographic population structure using genomic epidemiology approaches that employ bioinformatic and population genetic methods to better understand the evolution of these genes in the context of ancient diversity and human migration in this era of globalisation.
Research Group
Day Laboratory: Malaria genomics, epidemiology and control
Faculty Research Themes
School Research Themes
Key Contact
For further information about this research, please contact the research group leader.
Department / Centre
MDHS Research library
Explore by researcher, school, project or topic.