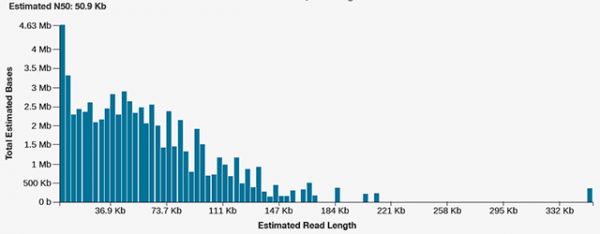
Nanopore GridION long-read sequencer
This resource is available to all University of Melbourne researchers and provides the opportunity to use the latest technology in long-read sequencing. We also provide flow cells for purchase at competitive prices in addition to GridION access.
About
Have you been thinking about utilising Nanopore sequencing for your research but don’t know where to start? Contact us to discuss using the GridION X5 long-read sequencer or potential collaborations.
The GridION offers real-time, long-read, high-fidelity DNA and RNA sequencing. Sequencing can be done on a wide range of starting samples including:
- Genomic DNA & cDNA – including targeted sequencing and detection of DNA modifications.
- Native RNA – including detection of RNA modifications.
- PCR amplified samples – ideal for limited sample input.
For further information please see: https://nanoporetech.com/products/gridion
The GridION platform is available to all University of Melbourne affiliated researchers and provides the opportunity to use the latest technology in long-read sequencing. We also provide flow cells for purchase at competitive prices in addition to access to the GridION instrument.
Prospective users will receive training in GridION use. Contact us for assistance with your long-read sequencing queries.
Contact
- GridION platform email & booking enquiries – long-reads@unimelb.edu.au
- Invoice enquiries: Franca Casagranda – f.casagranda@unimelb.edu.au
- Instrument expert & lab head: Dr Mike Clark – michael.clark@unimelb.edu.au
Location
PC2 Facility located in Room 2.56, Level 2, Melbourne Brain Centre (Building 144, Kenneth Myer Building, University of Melbourne, Royal Parade, Parkville).
Bookings
Available for booking for short (up to 4 hours) or long runs (up to 72 hours), with flow cell loading occurring during normal University of Melbourne opening hours Monday - Friday (0900 – 1700). Please note that outside of these hours, non-KMB staff will not be able to access the building or PC2 labs. Please contact us to be provided with a lab link to book GridION slots via Teamup. Current bookings can be viewed by following the link.
Getting started
- Data storage:
- If you are a Unimelb user make sure you have a Mediaflux project with at least 1 TB free.
- To request a Mediaflux project submit a Research Data Storage Request via this link http://go.unimelb.edu.au/z4q6
- For “Type of storage” select “Research data storage or access to data management platform” and in “ADDITIONAL INFORMATION” field say “Request for Mediaflux”
- To increase an allocation for an existing project http://go.unimelb.edu.au/zak6
- For “REQUEST TYPE” select “Research data management” and tell us which Mediaflux project you would like increased and by how much.
- To request a Mediaflux project submit a Research Data Storage Request via this link http://go.unimelb.edu.au/z4q6
- If you are not Unimelb user, please contact us to discuss possibilities.
- If you are a Unimelb user make sure you have a Mediaflux project with at least 1 TB free.
- Training:
- All users must be trained on flow cell loading and using the instrument prior to making a booking.
- Booking:
- All users must have a GridION slot booking before a sequencing run.
Data retention and clean-up policy
- If you are a Unimelb user all sequencing data can be uploaded from the GridION to your nominated destination Project on the MediaFlux server.
- You must have a MediaFlux account prior to performing your sequencing run with the recommended minimum storage space.
- If you are not Unimelb user, please contact us.
Data analysis
The GridION is managed and run by an academic group at the University and data analysis is not provided as a paid service. Interested in collaborating? Get in touch, we might be able to help.
Acknowledgements
Please acknowledge specific research staff, Dr Mike Clark, Dr Ricardo De Paoli-Iseppi or Franca Casagranda for their assistance during your experimental design or sequencing analysis process.